Efficacy of The SnotBuster™ (also known as SLSolution™) in Processing Lower Respiratory Tract Specimens for Routine Gram Stain and Culture
October 24, 2022
Key Insights:
- Study Validation: Researchers from the University of Florida College of Medicine validate The SnotBuster™ system (also known as SLSolution™) for processing lower respiratory tract specimens, comparing its performance against traditional routine methods.
- Performance and Sensitivity:The SnotBuster™ system demonstrated superior culture results, enabling work-up of significantly more pathogens on Day 1 compared to routine procedures, with enhanced ease of Gram stain interpretation and bacterial quantitation.
- Bacterial Growth and Morphology: The system allows for bacterial growth without inhibition, while maintaining clear bacterial morphology and cell structures, resulting in easier Gram stain readings and more consistent, reproducible specimen planting.
- Versatility in Clinical Settings: The SnotBuster™ system can be used in laboratories utilizing both manual and automated methods, offering greater quantities of organisms and enabling easier interpretation of Gram stains.
Estimated Reading Time: 8-10 Minutes
Forward
The data presented in this study were originally showcased at the American Society for Microbiology (ASM) Annual Meeting in 2021, one of the largest and most influential gatherings for microbiologists globally. This prestigious conference brings together researchers, clinicians, industry professionals, and thought leaders to share cutting-edge research, technological advancements, and practical innovations in the field of microbiology.
The ASM Annual Meeting provides a platform for presenting groundbreaking research across all branches of microbiology, including clinical diagnostics, microbial pathogenesis, and antimicrobial development. With a focus on fostering collaboration and the exchange of knowledge, the meeting is recognized as a key event for unveiling new technologies that can enhance laboratory practices and patient outcomes.
The SnotBuster™ system was featured in this forum to highlight its potential as a transformative tool in the processing of lower respiratory tract specimens, particularly in the context of diagnosing bacterial infections. By participating in this highly regarded event, the research team was able to demonstrate the system’s effectiveness to a wide audience of experts, gaining feedback and recognition from the microbiology community.
The study’s findings, adapted from the 2021 ASM poster presentation, underscore the significant impact that the SnotBuster™ system can have on clinical microbiology, offering improvements in bacterial growth, Gram stain clarity, culture work-up efficiency, and overall diagnostic accuracy.
Introduction
Culture and Gram stain (GS) of lower respiratory tract specimens (LRT) are routinely used to assist in determining the etiology of bacterial pneumonia. Appropriate specimen processing is key to the ultimate recovery of organisms in culture. LRT are often viscous, containing mucin molecules that trap microorganisms, causing them to clump together. Mucolytic agents, including dithiothreitol (DDT), are routinely used for processing specimens for mycobacteria smear and culture, but not for bacterial specimens in routine culture.
The purpose of this study was to compare the routine culture and GS results of LRT processed routinely (R) versus treatment with DDT in the SnotBuster™ system (SB) (Copan Diagnostics, Murietta, CA).
Materials and Methods
Specimen Processing
Lower respiratory tract specimens (sputum, endotracheal, or tracheal) were included in the study. Specimens were processed per routine laboratory protocol using a flocked swab for material selection for GS and culture onto sheep blood agar (BAP), chocolate agar (CHOC), and MacConkey agar (MAC).
The same specimens were processed using the SnotBuster™ system (Copan Diagnostics, Murietta, CA) within one hour of routine culture processing as follows:
- A Sputum Dipper (SD) was used to collect the respiratory specimen.
- The SD was placed in the Copan SL Solution test tube, capped, and vortexed for 30 seconds.
- The tube was incubated at room temperature for at least 15 minutes, but no longer than 60 minutes.
- The treated specimen was processed using a flocked swab for GS and cultured onto BAP, CHOC, and MAC.
Results
A total of 193 LRT were processed routinely and using The SnotBuster™ system.
Direct Gram Stain – Cells
- A total of 37/193 (19.2%) specimens were deemed poor quality (Q0) by both R-GS and SB-GS, not evaluated for bacteria, and not cultured.
- 3/193 (1.6%) were deemed Q0 by R-GS alone and were not cultured.
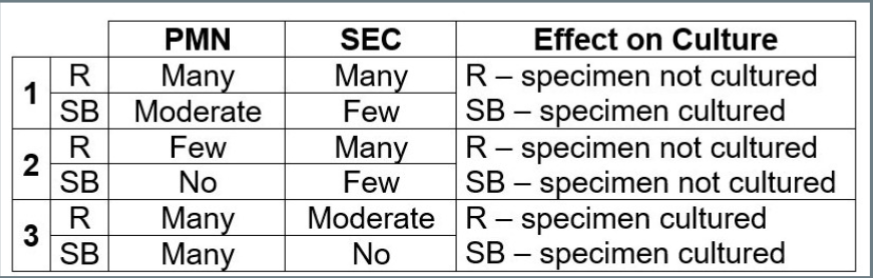
- There was an overall agreement of 98.4% (190/193) in the reporting of PMN and SEC between R-GS and SB-GS
Discordant results were as follows:
Culture Growth
153 cultures were evaluated:
- 3/153 (2%) cultures demonstrated no growth.
- 63/153 (41.2%) cultures grew Mixed Flora only:
- 48/63 (76.2%) grew the same amount in both R-C and SB-C.
- 9/63 (14.3%) grew significantly more in R-C than SB-C.
- 6/63 (9.5%) grew significantly more in SB-C than R-C.
Direct Gram Stain – Bacteria
153 cultures were evaluated:
- Bacteria were evaluated in 153 direct gram stains:
- 12/153 (7.8%) demonstrated no organisms.
- 115/153 demonstrated Mixed Flora:
- 114/115 (99.1%) showed agreement in reporting Mixed Flora between R-GS and SB-GS.
- 1/115 (0.9%) R-GS demonstrated significantly more Mixed Flora compared to SB-GS.
- 26/153 (17%) demonstrated a potential pathogen:
- 15/26 (57.7%) were reported in both R-GS and SB-GS.
- 6/26 (23%) were reported in R-GS only.
- 3/26 (11.5%) were reported in SB-GS only.
- 2/26 (7.7%) were overcalled in R-GS; an organism was called but did not grow in culture.
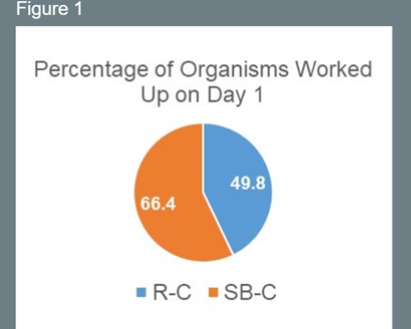
Culture Work Up on Day 1
153 cultures were evaluated:
- Bacteria were evaluated in 153 direct gram stains:
- 12/153 (7.8%) demonstrated no organisms.
- 115/153 demonstrated Mixed Flora:
- 114/115 (99.1%) showed agreement in reporting Mixed Flora between R-GS and SB-GS.
- 1/115 (0.9%) R-GS demonstrated significantly more Mixed Flora compared to SB-GS.
- 26/153 (17%) demonstrated a potential pathogen:
- 15/26 (57.7%) were reported in both R-GS and SB-GS.
- 6/26 (23%) were reported in R-GS only.
- 3/26 (11.5%) were reported in SB-GS only.
- 2/26 (7.7%) were overcalled in R-GS; an organism was called but did not grow in culture.
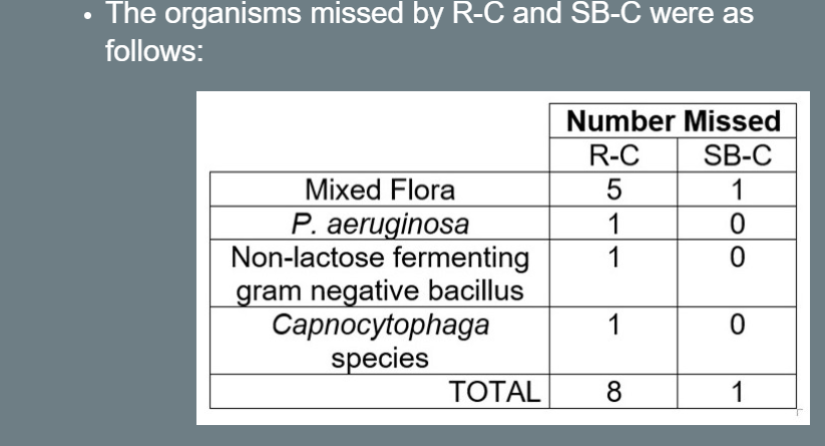
Missed Organisms
The organisms missed by R-C and SB-C were as follows:
- Mixed Flora: R-C (5), SB-C (1).
- P. aeruginosa: R-C (1), SB-C (0).
- Non-lactose fermenting gram-negative bacillus: R-C (1), SB-C (0).
- Capnocytophaga species: R-C (1), SB-C (0).
Day 1 Work-Up
- 87/153 (56.9%) cultures grew one or more potential pathogens ± Mixed Flora.
- 35/87 (40.2%) grew comparable quantities of organisms in both R-C and SB-C.
- 52/87 (59.8%) grew different quantities of organisms in R-C compared to SB-C.
- 18 cultures demonstrated greater quantities in R-C:
- 8/18 demonstrated greater quantities of a potential pathogen compared to SB-C.
- 10/18 demonstrated greater quantities of Mixed Flora but the same quantity of potential pathogen compared to SB-C.
- 44 cultures demonstrated greater quantities in SB-C:
- 29/44 demonstrated greater quantities of a potential pathogen compared to R-C.
- 15/44 demonstrated greater quantities of Mixed Flora but the same quantity of potential pathogen compared to R-C.
- 18 cultures demonstrated greater quantities in R-C:
Note: Lack of isolated colonies on Day 1.
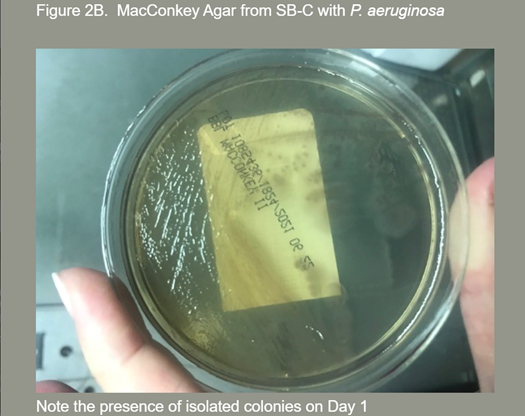
Note: Presence of isolated colonies on Day 1.
Sputum Dipper™ Transfer Device, Individually Packaged, Sterile
Discussion
- Quantitation of both cells and microorganisms was comparable between SB-GS and R-GS.
- SB GS was easier to interpret since both cells and microorganisms were more dispersed in the smear.
- Overall organism recovery was equivalent between SB-C and R-C. However, significantly more SB-C demonstrated greater quantities of organisms than R-C (See Figures 2A and 2B).
- Significantly more organisms were available for work up on Day 1 with SB-C compared with R-C.
Summary
The use of a mucolytic agent such as the SnotBuster™ system provides equivalent gram stain results and superior culture results, allowing for the work up of significantly more pathogens on Day 1 compared to routine procedures. This process can be used by clinical microbiology laboratories performing both manual and automated processing of lower respiratory tract specimens.
References:
- McCarter, Y. S., Reynolds, M., & Goglia, A. (2021). Efficacy of a novel device using dithiothreitol in processing lower respiratory tract specimens for routine Gram stain and culture. Poster presented at the American Society for Microbiology (ASM) Annual Meeting, Washington, DC.